qpAdm is a powerful tool used in population genetics to estimate the proportions of ancestry from different source populations in a target population. It is part of the ADMIXTOOLS software package developed by David Reich's lab at Harvard University. qpAdm is particularly important in studies of population history because it helps researchers understand the genetic contributions of various ancestral populations to modern groups.
Table of Contents
- Introduction
- What You Will Learn
- Assumptions
- Tools Used
- Understanding qpAdm Parameters
- Setting Up the Environment
- Preparing Your Data
- Running qpAdm Analysis
- Interpreting qpAdm Output
- Examples
- Troubleshooting and Tips
- Conclusion
- Glossary
Introduction
qpAdm is a powerful tool used in population genetics to estimate the proportions of ancestry from different source populations in a target population. It is part of the ADMIXTOOLS software package developed by David Reich's lab at Harvard University. qpAdm is particularly important in studies of population history because it helps researchers understand the genetic contributions of various ancestral populations to modern groups.
What You Will Learn
By the end of this tutorial, you will learn:
- The fundamental parameters and assumptions of qpAdm.
- How to set up the necessary software tools and prepare your data.
- The process of running a qpAdm analysis and interpreting the results.
- Common issues and best practices for qpAdm analysis.
Assumptions
This tutorial assumes that you have:
- Basic knowledge of Linux commands
Tools Used
Here are the tools and resources you will need for this tutorial:
- Ubuntu for Windows: Windows Subsystem for Linux (WSL) | Ubuntu
- AdmixTools by DReichLab: GitHub - DReichLab/AdmixTools: Tools test whether admixture occurred and more
- Additional details: Software | David Reich Lab (harvard.edu)
- Plink 1.90 (not 2.0): https://www.cog-genomics.org/plink/
- 23&me RAW DNA datafile
- AncestryDNA RAW DNA datafile
- Dataset: Allen Ancient DNA Resource (AADR): Downloadable genotypes of present-day and ancient DNA data | David Reich Lab (harvard.edu)
- Version v54.1.p1: 1240k (not 1240K + HO)
Understanding qpAdm Parameters
In qpAdm analysis, understanding the key parameters and their roles is essential for setting up your model accurately. qpAdm is a powerful statistical tool used in population genetics to estimate the proportions of ancestry from different source populations in a target population. Let's break down the main components.
Target Population
The target population is the group you are analyzing to understand its genetic ancestry. In this tutorial, we use "Target" as our target population label. The target population is typically the one for which you want to determine the proportion of ancestry from various source populations.
- The target is usually a present-day population or a recent historical population.
- You should have high-quality genetic data for your target population.
- The target population is assumed to be a mixture of the source populations.
Source Populations
Source populations are groups that you hypothesize have contributed to the ancestry of the target population. In qpAdm, you need to provide a list of at least two source populations. These populations should be genetically distinct and relevant to the historical context of the target population.
- Source populations should be older than or contemporaneous with the target population.
- They should represent potential ancestral populations based on archaeological, historical, or genetic evidence.
- The more distinct the source populations are from each other, the better qpAdm can distinguish their contributions.
For this tutorial, our example source populations are:
- Iran_ShahrISokhta_BA2: Representing ancient populations from Iran.
- Kazakhstan_Andronovo.SG: Reflecting ancient populations from Central Asia.
- Turkmenistan_Gonur_BA_1: Representing Bronze Age populations from Turkmenistan.
Right Populations (Right Pops)
Right populations, also known as right pops or outgroups, serve as reference points to help model the ancestry of the target population. They are crucial for distinguishing the ancestry contributions from different source populations. The right populations typically include a variety of ancient and modern populations that did not contribute directly to the target population. First population is always a basal African population, Mbuti.DG in this case. For this tutorial, our example right populations are:
- Mbuti.DG: An African basal population.
- China_Tianyuan: An ancient East Asian population.
- Karitiana.DG: A Native American population.
- Russia_Ust_Ishim_HG.DG: An ancient Siberian population.
- Ami.DG: A Taiwanese aboriginal population.
- Dai.DG: A Southeast Asian population.
- Turkey_N: An ancient population from Anatolia.
- Georgia_Kotias.SG: A prehistoric population from the Caucasus.
- Russia_Kostenki14.SG: An Upper Paleolithic European population.
- Iran_GanjDareh_N: An early Neolithic population from Iran.
Example of a Standard qpAdm Model
A standard qpAdm model can be expressed as follows:
Target population (Target) = source population 1 (Source 1) + source population 2 (Source 2)
The qpAdm output will provide:
- A p-value (also called tail probability or tailprob), which indicates the statistical fit of the model.
- Admixture coefficients (x and y) for Source 1 and Source 2, respectively, such that x + y = 1 (or 100%).
- Standard errors for the admixture coefficients, indicating the precision of the estimates.
Successful Model Criteria
For a qpAdm model to be considered successful, it should meet the following criteria:
- High p-value: Typically above 0.05, indicating a good fit between the model and the observed data.
- Low standard errors: Indicating precise estimates of the admixture coefficients.
- Positive admixture coefficients: Indicating plausible contributions from the source populations.
Setting Up the Environment
Before running qpAdm, you need to set up the necessary software tools and environment. This section will guide you through installing AdmixTools and setting up Plink.
Installing AdmixTools
- Download and extract AdmixTools:
- Visit the GitHub repository and download the latest version of AdmixTools.
- Extract the downloaded files to a directory on your computer.
- Navigate to the src/ directory:
cd AdmixTools-master/src - Install dependencies:
sudo apt-get install build-essential sudo apt-get install libgsl-dev sudo apt-get install libopenblas-dev - Compile the programs:
make clobber make all make install - Test qpAdm:
cd ../bin ./qpAdm
Setting Up Plink
- Download and extract Plink 1.90:
- Visit the PLINK website and download the latest version of Plink 1.90.
- Copy the executables:
- Extract the downloaded files and copy the Plink and prettify executables into the
binfolder of your AdmixTools directory.
- Extract the downloaded files and copy the Plink and prettify executables into the
Preparing Your Data
Proper data preparation is crucial for a successful qpAdm analysis. This section will guide you through downloading necessary datasets, processing raw data files, converting to EIGENSTRAT format, and merging with reference datasets.
Downloading Necessary Datasets
- Download the 1240k Eigenstrat database:
- Visit the Allen Ancient DNA Resource (AADR) and download the v54.1.p1_1240K_public dataset.
- Create a dataset folder:
- Create a new folder called
datasetin thebindirectory of your AdmixTools installation and copy the extracted dataset files into this folder.
- Create a new folder called
Processing 23andMe and AncestryDNA Files
Prepare 23andMe RAW Data File Using Plink
- Download and extract the 23andMe RAW DNA file:
- Follow the instructions on the PLINK website to download and extract your 23andMe raw data file.
- Convert the raw data file to binary format:
./plink --23file 23andme_Target_v5.txt Target 1 --make-bed --out Target_23andme_merged
Prepare AncestryDNA Data File
- Combine 23andMe and AncestryDNA raw data files:
- Refer to the guide on Genetic Lifehacks to combine your 23andMe and AncestryDNA raw data files.
- Strip header information from the AncestryDNA file:
awk 'BEGIN {FS="t"};{print $1"t"$2"t"$3"t"$4 $5}' AncestryDNA.txt > AncestryCombined.txt - Convert the processed file to binary format:
./plink --23file AncestryCombined.txt Target 1 --make-bed --out Target_Ancestry_merged
Converting to EIGENSTRAT Format
- Create a parameter file for conversion:
- Create a new parameter file called
convertf_param.parwith the following content:
genotypename: Target_23andme_merged_hh.bed snpname: Target_23andme_merged_hh.bim indivname: Target_23andme_merged_hh.fam outputformat: EIGENSTRAT genotypeoutname: Target_23andme_eigenstrat.geno snpoutname: Target_23andme_eigenstrat.snp indivoutname: Target_23andme_eigenstrat.ind - Create a new parameter file called
- Run the conversion:
convertf -p convertf_param.par
Merging with Reference Datasets
- Create a merge parameter file:
- Create a new parameter file called
merge_param.parwith the following details:
geno1: /path/to/AdmixTools/bin/dataset/v54.1.p1_1240K_public.geno snp1: /path/to/AdmixTools/bin/dataset/v54.1.p1_1240K_public.snp ind1: /path/to/AdmixTools/bin/dataset/v54.1.p1_1240K_public.ind geno2: /path/to/AdmixTools/bin/Target_23andme_eigenstrat.geno snp2: /path/to/AdmixTools/bin/Target_23andme_eigenstrat.snp ind2: /path/to/AdmixTools/bin/Target_23andme_eigenstrat.ind genooutfilename: /path/to/AdmixTools/bin/Target_merged_with_1240K.geno snpoutfilename: /path/to/AdmixTools/bin/Target_merged_with_1240K.snp indoutfilename: /path/to/AdmixTools/bin/Target_merged_with_1240K.ind outputformat: EIGENSTRAT - Create a new parameter file called
- Run the merge:
./mergeit -p merge_param.par
Running qpAdm Analysis
This section provides a detailed procedure for setting up and running qpAdm.
Creating Parameter Files
- Create folders for fstat results:
- Create new folders called
fstat_23andmeandfstat_ancestryin thebindirectory.
- Create new folders called
- Create the parameter file for qpfstats:
- Create a file called
parqpfstat.txtwith the following content:
DIR: /path/to/AdmixTools/bin S1: 10Oct21 S1X: 10Oct21 indivname: /path/to/AdmixTools/bin/Target_merged_with_1240K.ind snpname: /path/to/AdmixTools/bin/Target_merged_with_1240K.snp genotypename: /path/to/AdmixTools/bin/Target_merged_with_1240K.geno poplistname: /path/to/AdmixTools/bin/fstat_23andme/lista.txt fstatsoutname: /path/to/AdmixTools/bin/fstat_23andme/fstatsa.txt allsnps: YES inbreed: NO scale: NO - Create a file called
Setting Up Population Lists
- Create the population list file:
- Create a file called
lista.txtwith the following label names of each population:
Target Mbuti.DG Irula.DG Turkey_N Laos_LN_BA.SG China_Tianyuan Ami.DG Karitiana.DG Iran_GanjDareh_N Iran_C_SehGabi Iran_ShahrISokhta_BA1 Iran_ShahrISokhta_BA2 Turkmenistan_Gonur_BA_1 Dai.DG Russia_Ust_Ishim_HG.DG Chukchi.DG Saami.DG Georgia_Kotias.SG Russia_Kostenki14.SG Russia_Tyumen_HG Russia_MLBA_Sintashta Russia_DevilsCave_N.SG Luxembourg_Loschbour.DG Czech_BellBeaker Kazakhstan_Central_Saka.SG Portugal_MN.SG Kazakhstan_Andronovo.SG - Create a file called
Running qpfstats and qpAdm
- Run qpfstats:
./qpfstats -p fstat_23andme/parqpfstat.txt > fstat_23andme/qpfstatlog.txt ./qpfstats -p fstat_ancestry/parqpfstat.txt > fstat_ancestry/qpfstatlog.txt - Create the parameter file for qpAdm:
- Create a file called
parqpadm.txtwith the following details:
fstatsname: /path/to/AdmixTools/bin/fstat_ancestry/fstatsa.txt popleft: /path/to/AdmixTools/bin/fstat_ancestry/left.txt popright: /path/to/AdmixTools/bin/fstat_ancestry/right.txt details: YES - Create a file called
- Create the left and right population list files:
- Create
left.txtandright.txtfiles with the following content:
left.txt: Target Iran_ShahrISokhta_BA2 Kazakhstan_Andronovo.SG Turkmenistan_Gonur_BA_1right.txt: Mbuti.DG China_Tianyuan Karitiana.DG Russia_Ust_Ishim_HG.DG Ami.DG Dai.DG Turkey_N Georgia_Kotias.SG Russia_Kostenki14.SG Iran_GanjDareh_N - Create
- Run qpAdm:
qpAdm -p parqpadm.txt > qpadm_output.txt
Interpreting qpAdm Output
How to Read the qpAdm Output File
A successful qpAdm model will have:
- High p-value: Typically above 0.05, indicating a good fit between the model and the observed data.
- Low standard errors: Indicating precise estimates of the admixture coefficients.
- Positive admixture coefficients: Indicating plausible contributions from the source populations.
To diagnose why a model may have failed, refer to the generated Dstats, also known as gendstats in the output file. These statistics compare the simulated model to the actual target sample. Large Z scores (above 3 or below -3) can indicate why a model failed.
Examples
Using 23andMe Data
Using AncestryDNA Data
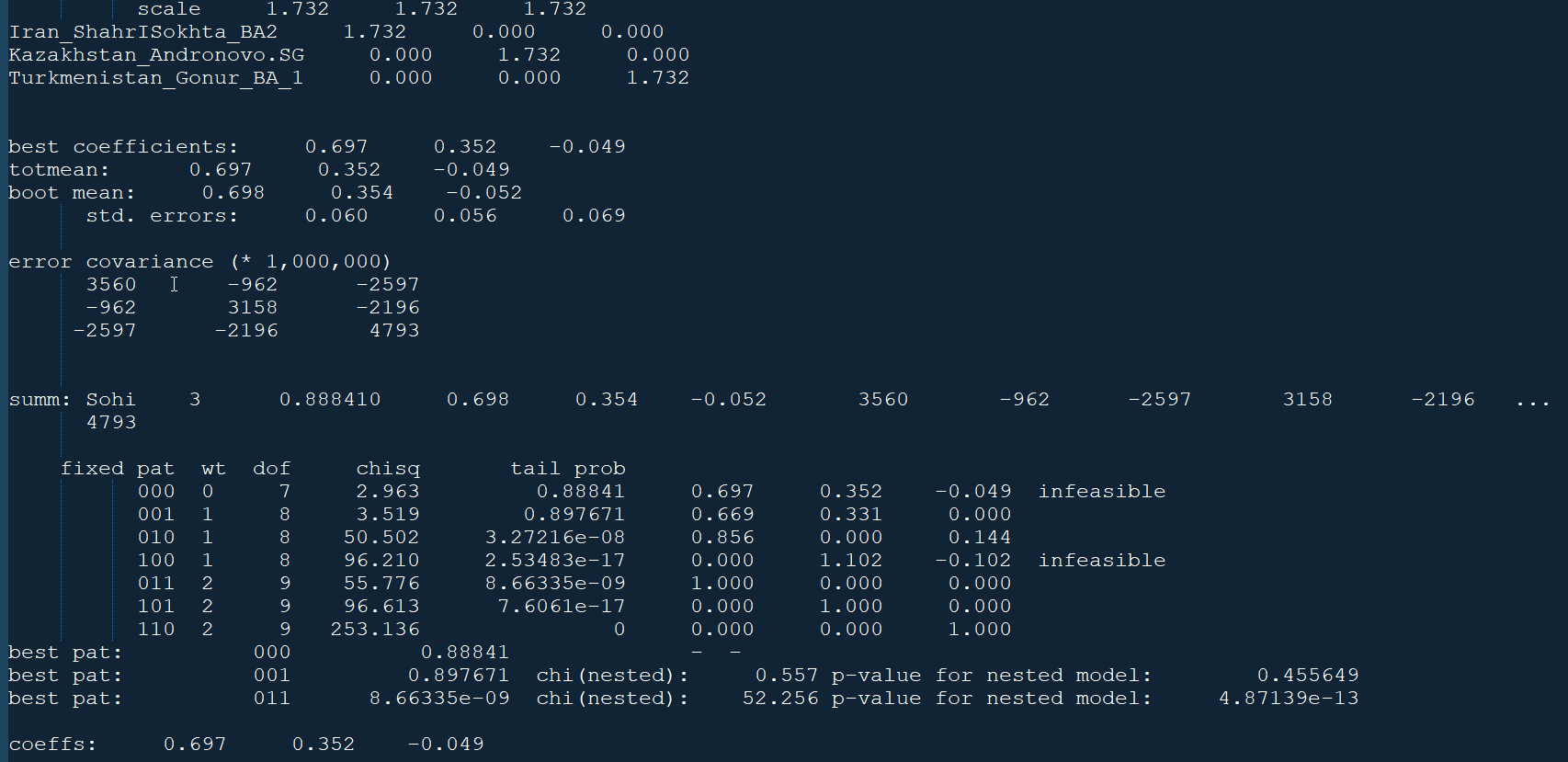
From the 23andMe output, our model with pattern 000 (i.e., all three populations) is infeasible even though the tail probability is 0.88841, which is > 0.05. This is because one of the admixture coefficients is negative. Pattern 001, meaning Iran_ShahrISokhta_BA2 and Kazakhstan_Andronovo.SG, seems to pass the test with a probability of 0.897671 and a coefficient of 66.9% for BA2 and 33.1% for Andronovo. Standard errors are also low at 0.06, 0.056, and 0.069.
Adjust the populations in left.txt and right.txt files and run the command again:
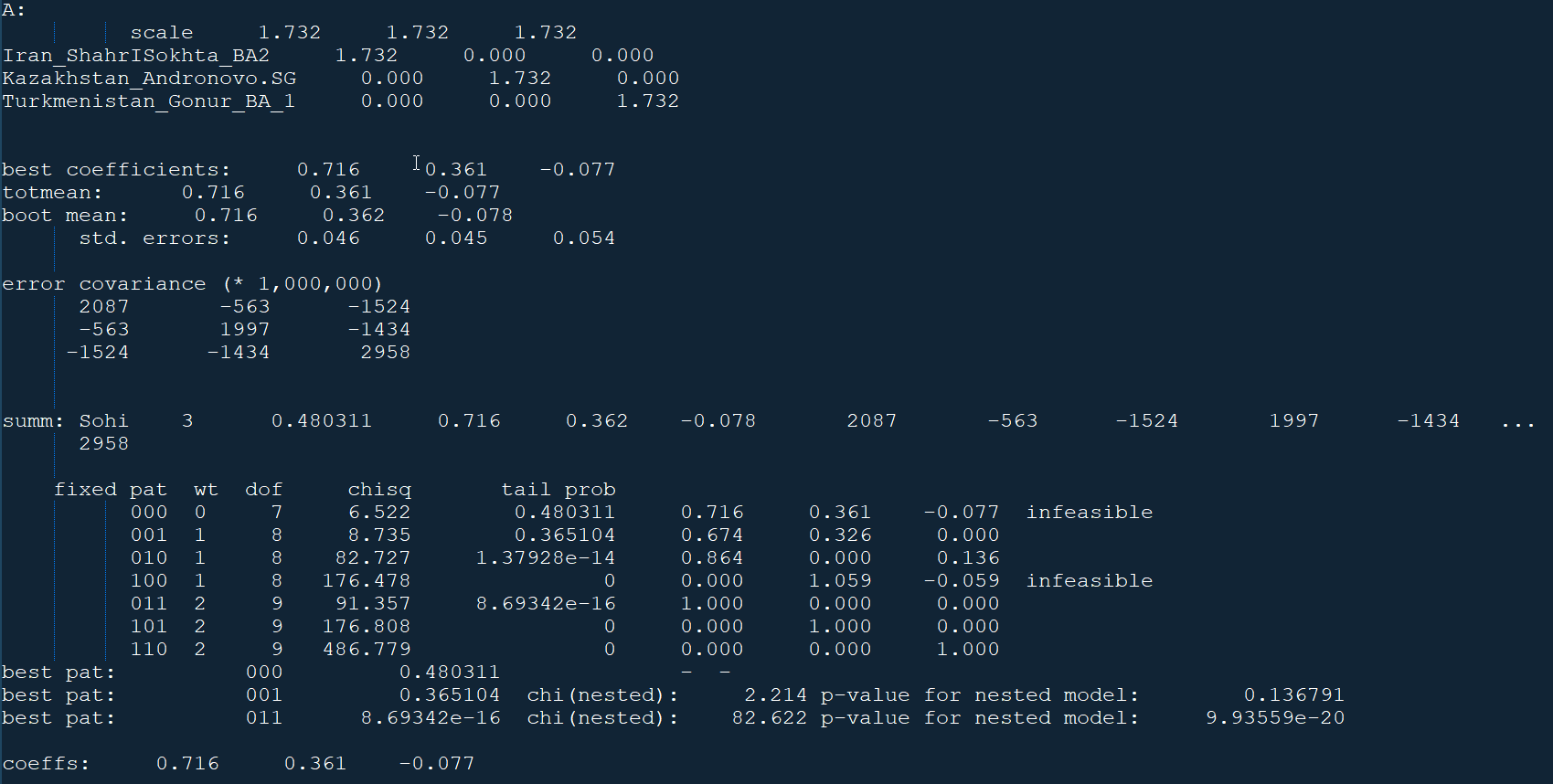
qpAdm -p parqpadm.txt > qpadm_output.txtWith the right combination of populations in left.txt, right.txt, and lista.txt, you can model your admixture. Example: qpAdm model output (sample)
Troubleshooting and Tips
Common Issues and Solutions
- Low p-value: Adjust the right populations to ensure they serve as proper outgroups.
- High standard errors: Ensure you have enough genetic data and that the populations are well-differentiated.
- Negative admixture coefficients: Check the left and right population lists for accuracy.
Best Practices for qpAdm Analysis
- Always verify your dataset for completeness.
- Use multiple outgroups to ensure robust results.
- Cross-validate with different datasets to confirm findings.
Conclusion
This tutorial has provided a comprehensive guide to using qpAdm for South Asian population models. By following the steps outlined, you can set up and run qpAdm analyses, interpret the results, and troubleshoot common issues. qpAdm is a powerful tool in population genetics, and mastering it can provide valuable insights into the ancestry of populations.
Glossary
- qpAdm: A tool used to estimate ancestry proportions from different source populations.
- ADMIXTOOLS: A software package for analyzing population genetics data.
- EIGENSTRAT: A data format used in population genetics.
- p-value: A measure of the validity of a model.
- Tail probability: Another term for p-value.
- Admixture coefficients: Proportions of ancestry from source populations.